Detection of Naturally Occurring Acyl- Homoserine Lactones Using a Chromobacterium Reporter Strain
Contributed by:
- John Lennox (Department of Microbiology, Penn State Altoona) Emeritus
- Bob McClean, Texas State University, San Marcos7
Instructor Version (go to Student Version)
| Subject Area(s) | Microbiology |
Intended Audience |
High school biology, independent study/science fair, introductory undergraduate microbiology, advanced college level microbiology. |
Type |
Laboratory exercise |
Revision Date |
May 5, 2010 |
CONTENT
Many functions in bacteria are regulated by a phenomenon variously called autoinduction or quorum sensing (QS). In QS, the bacteria constantly produce small highly diffusible molecules (often but not always acyl-homoserine actones [acyl-HSLs]), which cross the plasma membrane to the external environment. At low levels these molecules produce no evident effect. As population density increases or as diffusion is constrained, as in a biofilm for example, the concentration of A-HSL rises. At some threshold level the A-HSLs bind reversibly to specific m-RNA synthetase enzymes and these commence transcription of suites of genes previously inactive.
When first described as the mechanisms responsible for controlling bioluminescence Vibrio fisherii, autoinduction was thought a quaint if interesting phenomenon1. This view changed rapidly as A-HSLs were identified as the effector molecules2 and were shown to be functioning in a great number of additional bacterial systems including virulence3, motility4, pigment production in some bacteria5, and the develoment of normal biofilm architecture6, among many others.
It was not until 1997 that McClean et al.7 demonstrated the presence of A-HSLs in naturally occurring biofilms growing on limestone pebbles taken from the San Marcos river in Texas. There initial reporter system for A-HSLs was a strain of Agrobacterium tumefaciens bearing a lactose operon and lacking the ability to make its own A-HSLs. When a source of A-HSL of the appropriate type (N-oxo(octanoyl) HSL) was placed near the reporter strain on medium containing X-gal, a synthetic β-galactosidase substrate, the foreigh A-HSL induced the lac operon to synthesize β-galactosidase which in turn cleaved the X-gal turning the A. tumefaciens blue.

Although this represented a clever method of detecting naturally occurring A-HSLs, this system had some limitations as a teaching tool. The Lac operon was bourn on a plasmid that was unstable and had to be maintained by the inclusion of two antibiotics in the maintenance medium. The requirement for X-gal in the medium was an additional complication. Fortunately at the same time, another set of investigators had developed another reporter system using violacein production in Chromobacterium violaceum5.
C. violaceum strain CV026 fails to produce pigment due to the inability to produce the normal autoinducer N-hexanoyl HSL. Although normally non-pigmented, this strain responds to the presence of exogenous HSL by producing normal amounts of the violet pigment. C. violaceum strain 31532 over-produces the N-hexanoyl HSL, but fails to respond to it and is thus is also non-pigmented while the wild-type strain 12472 both produces and responds to N-hexanoyl HSL by producing violacein.
| Strain Designation | Description | Properties |
|---|---|---|
| ATCC 12472 | Wild Type (pigmented) | Produces N-hexanoyl-HSL (C6-HSL) and responds to same. |
| CV 026 | Reporter Strain (non-pigmented) | Fails to produce C6-HSL, but does respond to it. |
| ATCC 31532 | C6-HSL Producer (non-pigmented) | Over produces C6-HSL, but does not respond to it. |
These strains may be obtained from Robert McClean, Biology Department Texas State, San Marcos.
Safety Note: Chromobacterium violaceum is listed as a BSL 2 pathogen, in the same category with other organisms such as Klebsiella pneumoniae, Proteus vulgaris, Salmonella—all species, and Staphylococcus aureus. Instructors should consider this when deciding whether or not to adopt this exercise.
PREREQUISITES
Ideally this exercise should follow or be carried out in conjunction with a discussion of quorum sensing in the classroom. Students should be familiar with the basics of molecular biology, including transcription and translation.
INSTRUCTIONAL OBJECTIVE
The exercise described here can be used with entire classes in a manner similar to the original demonstration1, or it may be used as a jumping off point for students engaged in independent research. The exercise should reinforce the students understanding of gene induction, transcription, translation. The student may also observe that quorum sensing occurs in natural habitats and is not only a laboratory phenomenon. The techniques learned will provide the tools for students to conduct their own investigations in QS.
MATERIALS
| Quantity | Description |
|---|---|
| 1 culture | Chromobacterium violaceum Strain CV026 (reporter strain). |
| 1 culture | Chromobacterium violaceum Strain 31532 (over-producer of N-hexanoyl HSL). |
| 1 culture | Chromobacterium violaceum Strain 12472 (wildtype). |
| As Necessary | 3 plates of LB or R2A medium per student. |
| As Necessary | Source of biofilms (pebbles from streams, soil, aquarium samples, etc.). |
EQUIPMENT PER STUDENT
| Quantity | Description |
|---|---|
| 1 | Zip-loc plastic bag for collection samples |
| 1 | Sterile glass Petri dish |
| 1 | Pair forceps |
| 1 | Disinfectant for lab bench |
INSTRUCTIONAL PROCEDURES
Prior to the laboratory period but within 24 hours
- Prepare plates of LB or R2A medium according to the label instructions, 3 plates per student.
- Collect or have students collect and return to the laboratory samples suspected to contain naturally occurring biofilms, pebbles from streams, soil samples, scrapings from aquaria, material from sink traps, etc. Divide samples in two equivalent sets and autoclave one sample set to serve as a negative control. Alternatively, the rock can be sonicated and washed in sterile Distilled water to remove the biofilm.
During the Laboratory Period
- Have the students streak the three plates as shown in the following diagrams.
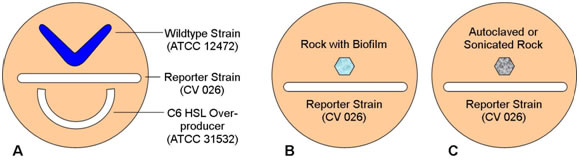 Figure 2.
Figure 2.
 Figure 3.
Figure 3.
- Remind students that strain 12472 is a wildtype which both produces and responds to N-hexanoyl HSL, strain CV026 responds to but does not produce N-hexanoyl HSL, and strain 31532 is an overproducer of N-hexanoyl HSL but does not respond to its presence. Have them predict the pattern of pigment formation in each plate.
- Incubate the plates at room temperature until the next lab period.
- Interpretation:
- Both the wildtype and the C6 HSL overproducer produce the HSL lacking in the reporter strain. CV 026 responds to both with the production of violacein
- B. If the rock sample has a biofilm component strain that produces N-hexanoyl HSL (C6-HSL) the reporter strain will produce pigment. Absence of pigment may indicate that a C6-HSL producer is lacking or that the rock was placed too far from the reporter strain on the plate.
- Heat killing or removal of the biofilm will remove C6-HSL and so the reporter strain should not react to the autoclaved or sonicated rock.
QUESTIONS
- Describe differences and similarities between Quorum Sensing as illustrated by pigment production in Chromobacterium violaceum and gene requlation as described in the Jacob and Monod model.
- If in your experiment with the rock containing a biofilm you observed no results, does this imply that no QS phenomena are taking place in the biofilm bacterial population? Explain.
- Spend a few minutes on the Internet and see how many other bacterial systems are known to be under the control of QS type systems in addition to pigment production.
ASSESSMENT / EVALUATION
Both students and instructors will be able to evaluate the effectiveness of the QS detection system by the response of the reporter strain in the control plate (A). A positive response in the “killed” plate (C) probably indicates an error in autoclaving or the student used the wrong strain as the “reporter”.
FOLLOW-UP ACTIVITIES
This assay system can be used as the starting point for any number of student generated research protocols.
- Individual bacterial strains isolated from nature can be tested for the production of C6-HSL.
- It has been demonstrated that other Acyl Homoserine lactones may interfere with the response to the native C6-HSL and cause a failure of pigment production in the wildtype strain (ATCC 12472). This phenomenon has been referred to as Quorum Sensing suppression. Bacterial strains and natural products (Herbs, spices, fruit juices, antimicrobials and many other materials can be tested for QS suppression.
- Ethyl acetate extracts of bacterial strains can be subjected to Thin Layer Chromatography and these TLC plates can then be overlaid with soft agar suspensions of the reporter strain to detect C6-HSL of the wild type strain, to detect other HSL secies by Quorum Sensing suppression.
- Another detection system using an Agrobacterium tumefaciens strain bearing a Lac operon can be used to detect a wider range of Acyl-HSLs. (See McClean R.J.C. et. al, 1997).
REFERENCES
- Nealson, K.H., Platt, T. and Hastings, J.W., 1970. Cellular Control of the Synthesis and Activity of the Bacterial Luminescent System. Journal of Bacteriology 104: 313-322.
- Fuqua, W.C., Winans, E.P., and Greenberg, E.P. 1994. Quorum Sensing in Bacteria: the Lux R-Lux I Family of Cell Density-Responsive Transcriptional Regulators. Journal of Bacteriology 176: 269-275.
- Passador, L., Gambello, M.J., Rust, L., and Iglewski, B.H. 1993. Expression of Pseudomonas aeruginosa Virulence Genes Requires Cell-to-cell Comunication. Science 260: 1127-1130.
- Hentzer, M., Givskov, M., and Eberl, L. 2004. Quorum Sensing In Biofilms: Gossip in Slime City. IN Microbial Biofilms, Ghannoum, M., and O’Toole, G.A., Eds.
- McClean, K.H., Winson, M.K., Fish, L., Taylor, A. Chhabra, S.R., Daykin, M., Lamb, J.H., Swift, S., Bycroft, B.W., Stewart, G.S.A.B., and Williams, P., 1997. Quorum Sensing and Chromobacterium violaceum: Exploitation of Violacein Production and Inhibition for the Detection of N-acylhomoserine lactones. Microbiology 143: 3703-3711.
- Davies, D.G. Parsek, M.R., Pearson, J.P., Iglewski, B.H., Costerton, J.W., and Greenberg, E.P. 1998. The Iivolvement of Cell-to-cell Signals in the Development of a Bacterial Biofilm. Science 280: 295-298.
- McClean, R.J.C., Whiteley, M., Stickler, D.J., and Fuqua, W.C., 1997. Evidence of Autoinducer Activity in Naturally Occurring Biofilms, FEMS Microbiology Letters 154: 259-263.
This material is based upon work supported by the National Science Foundation under Grant No. 0618744. Developed in collaboration with Dr. John Lennox, Penn State Altoona. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
©2002-2008 Center for Biofilm Engineering, http://www.biofilm.montana.edu
