Genes and Biofilm Formation
Are the conditions governing biofilm formation stochastic and random such as the hydrodynamic forces or nutritional availability considered above, or are they deterministic, that is, determined by the organisms themselves?
Only a few years ago, it was thought that physical factors such as hydrodynamics and perhaps nutrition were the sole or at least dominant forces determining the structure of biofilms. Now, following a brilliant series of experiments in laboratories around the world it has been determined that the genomes of biofilm constituent organisms play a significant, perhaps a primary, role in biofilm formation. Using various forms of mutagenesis including chemical mutagens and transposon knock out techniques and an assay for biofilm production using 96 well plates a large number of mutants, generally called sad (surface association deficient) mutants, microbiologists have been begun to genetically dissect the pathway to biofilm formation in several important pathogens including, E. coli, Pseudomonas aeruginosa and Vibrio cholera. Sad mutants have been found which affect major steps in the developmental pathway of biofilm development.
The Assay for biofilm deficient mutants
The first sad mutants were isolated in the laboratories of George O’Toole and Roberto Kolter at the Dartmouth and Harvard Medical Schools respectively. The technique used to identify them had been developed with this goal in mind. Ninety-six well polyvinylchloride (PVC) plates with nutrient medium, are inoculated with various strains of microorganisms (various presumptive sad mutants for example). The plates are rocked or gently shaken to permit growth and encourage biofilm formation. After an incubation period of 8 hours, the plates are washed with water to remove any planktonic cells. Crystal violet (or Safranin) dye is then added to the wells to stain any biofilm adhering to the polystyrene walls of the well plate. The plates are then again washed thoroughly to remove any unbound crystal violet dye. Alcohol is then added to the wells in order to elute the crystal violet dye adhering to the biofilm. The amount of dye that dissolves in the alcohol is proportional to the amount of biofilm adhering to the walls of the plate. Wells containing sad mutants will be notably free of crystal violet dye at this point (See Exercise 6, Colorimetric Measurement of Biofilm Density).
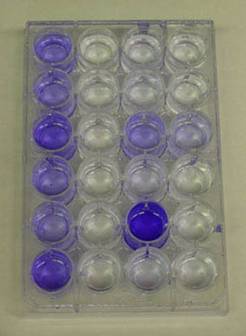
Some ten years ago (1998) O’Toole and Kolter reported observations on some of their first sad mutants. These biofilm deficient strains lacked either the polar flagellae possessed by normal Pseudomonas aeruginosa strains or lacked the Type IV pili common in the same organism. What did the lack of these two organelles, both associated with motility, have to do with biofilm formation?
Bacterial flagellae are coiled tubular structures made of protein (flagellin) and rotated by a complex motor consisting of several parts and powered by a proton motive force generated at the cytoplasm cell membrane interface. The effect of the rotation of this coiled structure is to propel the bacterium through its medium (water, blood etc.) at rates of up to 60 cell lengths sec -1
The wild type strain of P. aeruginosa forms a mono-layer of cells attached to the PVC plate within 8 hours. A mutant strain, however, carrying a defect in the flgK gene determining the loss of the ability to produce flagellae exhibited a significant reduction in attachment. The authors speculate that the flagellum is a significant factor in overcoming repulsive forces acting at the hydrodynamic boundary layer and thus, without the propulsive force of the flagellum, fewer bacteria come in contact with the surface.
Click image shown above to view video, which is included in the web page holding the Hoch-Burr Lab Xylella fastidiosa movies
Cornell University)
Type IV pili are involved in a curious type of motion called “twitching motility”. In the wild type P. aeruginosa, once cells have attached reversibly to the PVC plate, they can be seen to move in a rapid twitching manner to form micro-colonies with nascent channels in between. These microcolonies are the first indication of organization and “structure” in the developing biofilm. The Type IV pilus mutant was perfectly capable of forming the initial monolayer of cells, but lacked the ability to form micro-colonies due, presumably, to the inability to perform “twitching motility” resulting in a flat, dense, unstructured biofilm lacking microcolonies and water channels. This indicates a role for Type IV “twitching motility” in the early organization of biofilm structure (O’Toole and Kolter 1998).
Other contact related bacterial structures associated with biofilm formation have been recognized, including the curli fimbriae and Type I pili in E. coli, mutants deficient in flagellae, type IV pili and EPS in V. cholera and many others.
Paul Kolenbrander’s group at the NIH have reported a complex set of protein carbohydrate binding mechanisms among the hundreds of bacteria that inhabit the teeth in humans. These adhesins permit the “coaggregation” of bacteria in successional pattern leading to the formation of intricately structured biofilms in the plaque of human subjects. The genes related to many of these adhesins are now known and these control the sequence of binding of bacteria to the growing biofilm.
In fact the array of different structures found to be necessary for initial attachment is very large and so diverse that the overlap of genes from one organism to another seems to be surprisingly small.
It well may be the case that even within a given organisms there are multiple pathways to the production of biofilms each keyed to a unique carbon source. O’Toole et al. (1998), showed that a biofilm deficient mutant originally selected on supplemental amino acids could be completely restored to normal biofilm phenotype by changing the carbon source to citrate or glutamate. The authors interpreted this observation to independent, convergent pathways to biofilm formation triggered by different nutrient sources.
Mutants have been identified that are responsible for the growth and maturation of biofilms. In P. aeruginosa, for example, contact with a surface has been shown to be associated with up-regulation of the algC gene, which codes for an enzyme controlling the synthesis of alginate. This polysaccharide is the most abundant extracellular material in the matrix of P. aeruginosa. Simultaneously with the induction of algC, genes for the production of flagellae are down regulated. These two functions, one associated with planktonic life and the other associated with the biofilm phenotype are co-regulated by a common gene expression factor called s22, as one is turned on the other is turned off. A very similar situation has been observed in E. coli with the up-regulation of cholonic acid a major matrix exopolysaccharide with the down-regulation of flcC, which codes for the flagellar structural protein.
Similar sad mutants in other organisms have been shown to be deficient in other matrix and cell surface components including the lipopolysaccharide (LPS) of the gram-negative cell wall.
Signaling and Quorum Sensing
In 1997, McLean et al. carried out an elegantly simple experiment demonstrating that QS is more than a laboratory phenomenon. The study employed Agrobacterium tumefaciens indicator strains unable to make their own signaling molecules but which possessed the ability to respond to exogenous autoinducers. The indicator strains had a Lac (lactose) operon down stream from the promoter normally activated by the signaling molecule. These indicator strains were streaked onto plates of medium containing a lactose analog (X-gal) and nearby (about 1-2 cm) was placed a pebble isolated from a local stream. The pebble was covered with a naturally occurring slimy biofilm.
As the indicator strain grew on the medium those portions of the streak closest to the pebble turned blue as a result of the activation of the promoter region and the down stream lac operon. b-galactosidase a product of the induced lac operon cleaves the X-gal substrate releasing a blue colored product. This simple demonstration was the first indicating the action of QS systems and acyl-homoserine lactones in a natural biofilm.
Image of McLean et al (McLean, R.J.C., Whiteley, M., Stickler, D.J., and Fuqua, W.C.(1997) Evidence of autoinducer activity in naturally occurring biofilms. FEMS Microbiol Letts 154: 259–263.) Image showing HSLs on pebbles.
Subsequently Charlton et al. 2000 demonstrated why it is that the Marine Algae Delesia pulchera, unlike most other marine algae, Delesia pulchera grows free from algal contamination. Delesia synthesizes several members of a family of compounds called Furanones, which are naturally occurring structural analogs of the acyl-homoserine lactones responsible for most QS activity in gram-negative bacteria.
Although acyl-HSLs are present in biofilms many wondered if their presence was a primary factor, necessary for biofilm development of a secondary one, a consequence of prior biofilm formation. A study by (Davies et al. 1998) has pointed to the first hypothesis. Mutants of Pseudomonas aeruginosa (Las-I gene mutations) deficient in the ability to produce N-(3-oxododecanoyl)-L-homoserine lactone were unable to form the normal three dimensional tower and mushroom-like biofilms typical of this species. Instead, the biofilms formed were denser, and only about 20% the thickness of the wild type biofilms and lacked the water filled channels usually found in Pseudomonas biofilms. These biofilms were, in fact, quite similar to those produced by mutant strains lacking the Type IV pilus. Addition of the missing LasI gene product (N-(3-oxododecanoyl)-L-homoserine) to the growth medium restored the normal architecture of the wild type strain with respect to cell density and biofilm thickness. Davies et al. also observed that this QS mutant lacked one of the other properties usually noted in biofilms in that the lasI mutant was sensitive to the biocide sodium dodecyl sulfate that normal biofilms resist.
Many reports have demonstrated the presence of acyl HSLmolecules at biologically effective levels (nanomolar) in the lungs and sputum of cystic fibrosis patients.
These experiments supports the hypothesis that QS is essential for the normal development, if not the initiation, of biofilm development.
Following the initial observations of the role of quorum sensing molecules (acyl-HSLs) in the maturation of biofilms there have been intensive studies in many laboratories of similar phenomena and it now appears clear that upon biofilm formation in E. coli, for example, as much as 40% of the genome may be up or down regulated by at least a factor of 2 fold. Lejeune (2003) gives a partial list of genes differentially expressed in planktonic cells and those in biofilms on abiotic surfaces and the list includes genes for exo and lipo pollysaccharides, motility and attachment, proteins involved in secretion and transport, in carbon and nitrogen metabolism in transcription and translation and in drug resistance. There will likely be grist for many, many Masters theses and PhD dissertations before this molecular jungle is sorted out.
References
Davies, D.G., M.R. Parsec, J.P. Pearson, B.H. Iglewski, J.W. Costerton and E.P. Greenberg, 1998. The Involvement of Cell-o-cell Signals in the Development of a Bacterial Biofilm, Science 280: 295-298.
O’Toole, George A., Leslie A. Pratt, Paula I. Watnick, Dianne K. Newman, Valerie B. Weaver and Roberto Kolter, 1999, Genetic Approaches to Study.
Biofilms, IN, Methods in Enzymology: Volume 310, pp. 91-109, Ron J. Doyle Ed. Academic Press, San Diego.
O’Toole, G. A., and R. Kolter, 1998. Flagellar and Twitching Motility are Necessary for Pseudomonas aeruginosa Biofilm Formation, Molecular Microbiology 30: 295-304.



